Single-cell metabolomics offers a powerful approach for characterizing metabolic heterogeneity at the individual-cell level that is obscured in bulk analyses. Mass spectrometry-based single-cell metabolomics enables the label-free, high-throughput detection of metabolites; however, current methods are not sensitive enough to detect low-abundance metabolites, and they show high technical variability and restricted coverage and annotation confidence. In a study published in Nature Methods, research teams led by Prof. ZHU Zheng-Jiang at Shanghai Institute of Organic Chemistry (SIOC) of the Chinese Academy of Sciences develop an ion mobility-resolved mass cytometry technology for single-cell metabolomics . By integrating novel data acquisition with advanced processing strategies, this single-cell metabolomics method achieves high throughput while maintaining deep coverage. Researchers first established an ion mobility-resolved mass cytometry platform by integrating flow cytometry with ion mobility–mass spectrometry (IM–MS). The system continuously injects live cells into the IM–MS for direct electrospray ionization, transfers the pulsed single-cell ions that are generated into an ion trap for accumulation, and performs ion mobility separation with multidimensional measurements. To improve the detection of low-mass metabolites and reduce interference from cellular lipids, researchers implemented selective ion accumulation in the low-mass range on trapped ion mobility spectrometry. With this strategy, the abundance of low-mass ions was enhanced, increasing the sensitivity of detection for low-mass metabolites by 20-fold compared to the default condition. In addition, to address the low robustness of low-mass metabolites, a cell superposition strategy was developed. This strategy leverages distinctive ion signatures characterized by mass-to-charge ratio (m/z) and ion mobility values to aggregate identical ions across multiple cells. It enhanced peak detection and guided targeted ion extraction in individual cells, thereby improving robustness while maintaining single-cell resolution. This strategy elevated the median signal-to-noise ratio by 33-fold in peak detection and reduced metabolite dropout rates from 82% to 6% in quantification. Together, the selective ion accumulation and cell superposition enabled attomole-level sensitivity and a broad dynamic range at the single-cell level. Then, researcher developed MetCell, an end-to-end computational tool optimized for ion mobility-resolved single-cell metabolomics. Overall, the technology detected over 5,000 metabolic peaks and annotated approximately 800 metabolites per cell, representing a 3–10-fold improvement over existing approaches. Notably, 389 metabolites were identified with level 1 confidence, which is the highest number reported at this level in single-cell metabolomics currently reported. Finally, using this technology, researchers curated a metabolic single-cell atlas containing 45,603 primary liver cells from aging mice. They demonstrated accurate cell type and subtype annotation using single-cell metabolomics and unveiled distinct metabolic states and heterogeneity of hepatocytes during aging.
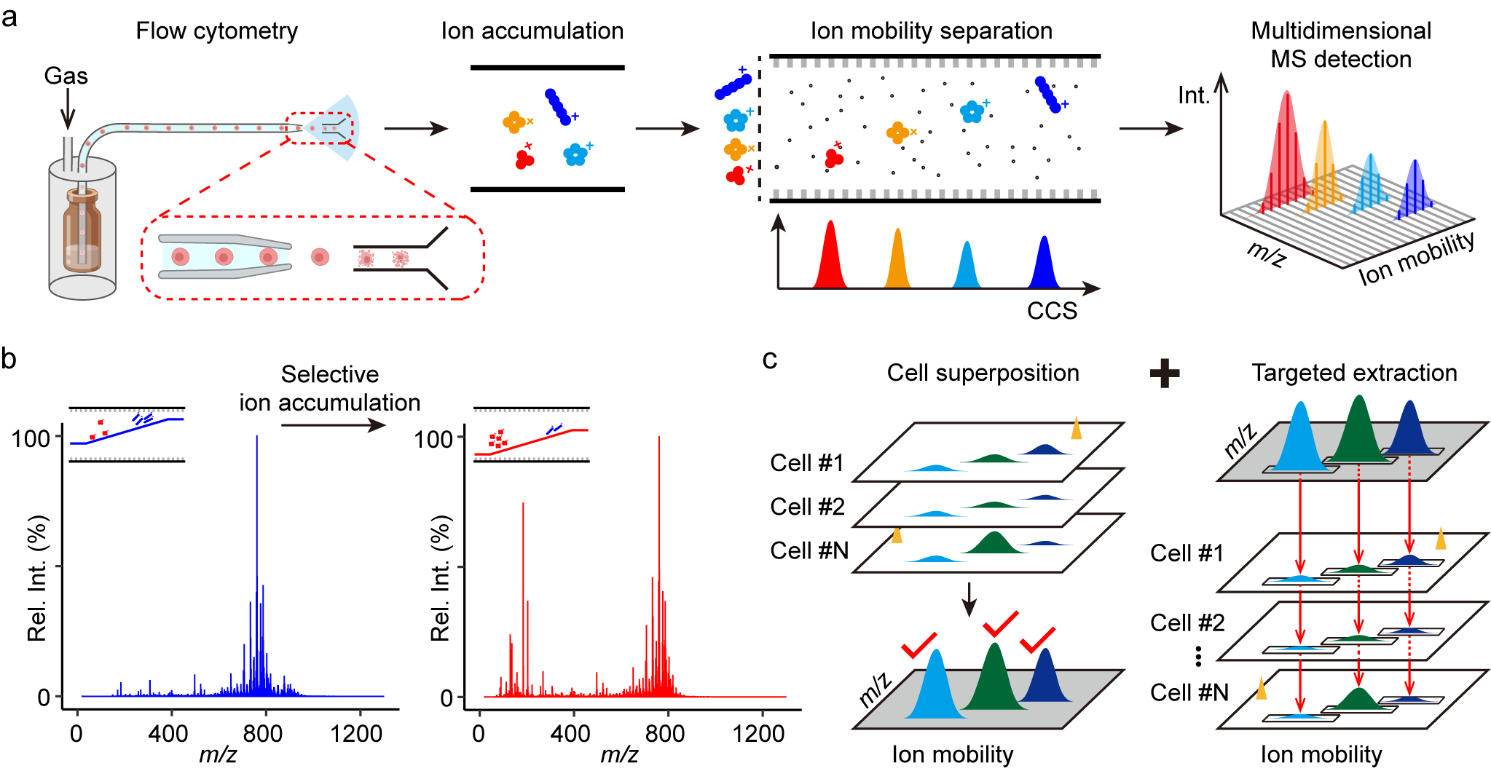
Figure 1. Ion mobility-resolved mass cytometry for single-cell metabolomics. (Image by ZHU Zheng-Jiang)
Professor ZHU Zheng-Jiang Interdisciplinary Research Center on Biology and Chemistry, Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences 100 Haike Road, Building 13, Pudong, Shanghai, P. R. China 201210 Tel: 0086-21-68582296 Email: jiangzhu@sioc.ac.cn
|


