Ubiquitination is a prevalent form of post-translational protein modification in mammalian cells, primarily including mono-ubiquitination and poly-ubiquitination with different linkage types. Through its unique molecular mechanisms, this important modification plays a central regulatory role in various key cellular biological processes. Among them, K48-linked ubiquitin chains mainly participate in the ubiquitin-proteasome degradation pathway, responsible for intracellular protein quality control. Shigella flexneri is a common pathogenic bacterium that secretes multiple effector proteins during host cell invasion to promote its survival and proliferation within host cells. The IpaH1.4 effector protein is a NEL-type E3 ubiquitin ligase secreted by S. flexneri, which can suppress host innate immune responses by mediating K48-linked ubiquitination and proteasomal degradation of relevant host proteins. The research team led by Prof. PAN Lifeng at the State Key Laboratory of Chemical Biology, Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences, has long been dedicated to studying the mechanism of the effector protein IpaH1.4. The team's previous research study revealed, for the first time, the mechanism by which the effector protein IpaH1.4 employ various "tricks" to disrupt the linear ubiquitin chain assembly complex (LUBAC) (PNAS. 2022, 119(12), e2116776119). However, other potential human host proteins targeted by IpaH1.4 and their related molecular mechanisms remain not well understood. Recently, in a collaborative study published in Nature Communications titled "Shigella effector IpaH1.4 subverts host E3 ligase RNF213 to evade antibacterial immunity", the research teams of Prof. Lifeng Pan and Yixiao Zhang from Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences, and Yufeng Yao from Shanghai Jiao Tong University School of Medicine made new discoveries. In this study, the team members firstly identified through affinity mass spectrometry that the key immune factor E3 ubiquitin ligase RNF213 is another important substrate protein of IpaH1.4 in human host cells. Notably, RNF213 is the largest E3 ubiquitin ligase in human cells, consisting of 5207 amino acid residues, and its genetic variation can cause cerebrovascular abnormalities and lead to Moyamoya disease. Subsequently, through systematic biochemical, structural biology, and cell biology analyses, they revealed, for the first time, that the E3 effector protein IpaH1.4 of S. flexneri can directly target human RNF213 via a specific interaction between its LRR domain and the RING domain of RNF213, and mediate K48-linked ubiquitination and proteasomal degradation of RNF213 in cells. Furthermore, the team members determined the cryo-EM structure of human RNF213 and the crystal structure of the IpaH1.4 LRR/RNF213 RING complex, elucidating the molecular mechanism underlying the specific recognition of RNF213 by IpaH1.4. Finally, relevant cell-based functional assays demonstrated that the targeting of host RNF213 by IpaH1.4 promotes the proliferation of S. flexneri within infected cells. 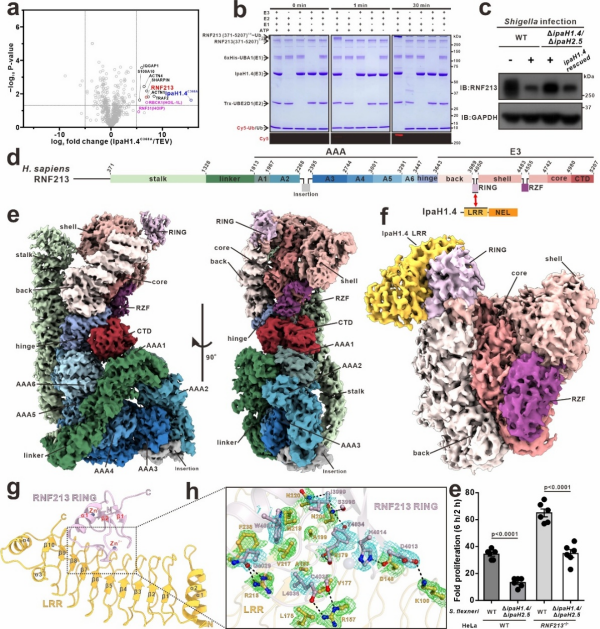
Figure 1. The molecular mechanism of Shigella flexneri E3 ubiquitin ligase effector IpaH1.4 in counteracting host immune protein RNF213 Finally, based on our findings in this work and previous reports, the team members proposed a model depicting the targeting and subversion of host RNF213 by S. flexneri effector IpaH1.4 as well as the counteraction of S. flexneri with the relevant RNF213-mediated host antibacterial processes during invasion. In this model, once S. flexneri invades the cytosol of a host cell, its LPS molecules on the outer membrane will be specifically recognized by RNF213, which can further mediate the ubiquitylation of LPS and in turn recruit LUBAC to mediate the linear ubiquitination on the invading bacterial surface. Then, relevant downstream linear ubiquitin-binding proteins, such as NEMO and Optineurin, will be recruited to trigger inflammation and antibacterial selective autophagy processes to suppress the replication of invading bacteria in the host cell. However, S. flexneri can secrete the E3 effector IpaH1.4 to directly target and hijack RNF213 via a specific interaction between IpaH1.4 LRR and RNF213 RING uncovered in this study. Subsequently, IpaH1.4 can mediate the K48-linked ubiquitination and proteasomal degradation of RNF213, thereby counteracting the relevant host antibacterial immune processes induced by RNF213 to facilitate the proliferation of S. flexneri in host cells. 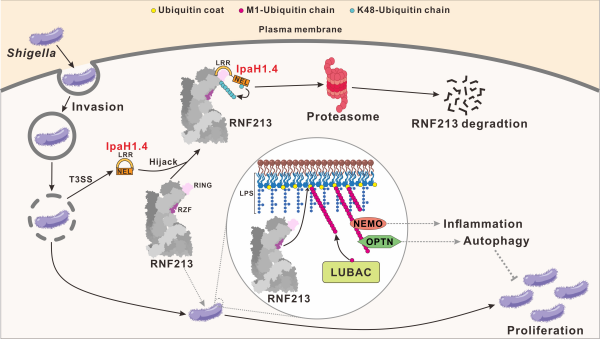
Figure 2. Schematic model of Shigella flexneri counteracting host RNF213 through E3 effector proteins IpaH1.4. The PhD students Xindi Zhou, Huijing Zhang and the post-doctoral fellow Yaru Wang from Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences, as well as associate professor Danni Wang from Shanghai Jiao Tong University School of Medicine are the co-first-authors of this paper. The corresponding authors are: Prof. Lifeng Pan and Prof. Yixiao Zhang from Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences; and Prof. Yufeng Yao from Shanghai Jiao Tong University School of Medicine. This work was supported in part by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences Grant (No. XDB1060000 for L.P.), National Natural Science Foundation of China (32071219, 92253301, 21822705 for L.P., 81830068 for Y-F.Y., 82472285 for D.W., and 32071297 for J.L.), the National Basic Research Program of China (2022YFC2303102 for L.P.), the CAS Youth Interdisciplinary Team (JCTD-2022-10 for L.P.), the STI2030-Major Projects (2022ZD0207400 for Y.Z.), Shanghai Key Laboratory of Aging Studies (19DZ2260400 for Y.Z.), the Basic Research Pioneer Project by the Science and Technology Commission of Shanghai Municipality (STCSM for Y.Z.), and the Foundation of Key Laboratory of Veterinary Biotechnology, Shanghai, P.R. China (No. BD1500010 for D.W). |


